
Introduction
Achieving a successful crop relies on a delicate balance between the use of available resources and avoidance of risks, resulting in a maximized economic output. For cereals, this output is grain yield, produced in inflorescences at the end of their annual cycle. Risks (frost, heat, pests, diseases, etc.) are not equally probable throughout the growth cycle of the plant, and neither is the availability of water and nutrients. Likewise, the plant is not equally susceptible to risks, nor is it equally sensitive to lack of resources at each growth phase. For a cereal like barley (Hordeum vulgare L.), flowering time is the key factor for adaptation and can be partitioned into two major developmental phases, in which different yield components are determined: (i) time from sowing to appearance of first stem node (containing vegetative and early reproductive phases), in which leaves and spikelets are formed, and (ii) late reproductive (Slafer and Rawson, 1994), in which floret development takes place. The morphological milestones plant emergence, appearance of the first node, and heading time, set the limits for these phases. Barley phasic development is controlled by an interaction of genetic and environmental factors. The main environmental factors are photoperiod and temperature (Laurie et al., 2004). On the other hand, allelic combinations at key loci determine the onset and duration of developmental phases, affecting plant resource allocation and source-sink balance. Adaptation to a specific environment is achieved by selection of the best combinations of alleles that optimize yield at each specific environment.
Barley (like wheat) can be sown in autumn or spring. Barleys adapted to autumn sowing are said to have a “winter growth habit,” whereas those sown in spring display a “spring growth habit,” the growth habit corresponding to the set of genetic features making the crop particularly fitting to each sowing date. Winter type-barleys require a period of low temperatures (vernalization) to accelerate flowering (while contributing to withstand frosts), whilst spring barleys grow in absence of cold temperatures (Trevaskis et al., 2003).
Although barley is a crop exhibiting one of the widest distributions across agroecological zones, current adaptation syndromes may not be optimum for some of the new conditions generated by climate change, posing an extra burden on plant breeders. New cultivars should be prepared to withstand and even thrive in the new climatic conditions. Under these circumstances, it seems sensible to explore and test new allelic combinations (or at least new to a particular region) of the key development genes that may perform better than the current ones.
Among the changes expected in climate, there is wide consensus on the increase of daily and night ambient temperatures (IPCC, 2013). This increase may affect barley growth in different ways. On the one hand, it may compromise fulfilling the period of cold temperature required by winter-type barleys to complete vernalization. On the other hand, higher ambient temperatures are predicted to hasten growth (Asseng et al., 2015) and may affect gamete production and spike fertility (Prasad and Djanaguiraman, 2014). It has been reported that warmer night temperatures during the critical period, comprising from the third detectable node to 10 days after anthesis, caused a shorter grain filling with negative consequences on grain yield, associated to decrease of grain number (García et al., 2015, 2016; Giménez et al., 2021). It is crucial to explore crop developmental responses to climate change. Particularly, it is important to understand how elevated temperature affects flowering time and its components: (i) duration of pre-flowering phases, (ii) number of leaves initiated in main shoots and their rate of appearance (or its inverse, phyllochron, defined as thermal time between the emergence of two successive leaves) (Slafer and Rawson, 1994; Jamieson et al., 1998). Likewise, it is essential to analyze how genetic variation in genes regulating flowering time influences or modifies the responses to high temperatures (Kiss et al., 2017).
Vernalization requirement in barley is controlled by three loci: VRN-H1, VRN-H2 and VRN-H3, located on chromosomes 5H, 4H and 7H, respectively. VRN-H1 is a flowering inducer gene that encodes an AP1-like MADS-box transcription factor (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). VRN-H2 is a flowering repressor, encoded by a zinc finger-CCT (CONSTANS, CONSTANS-like, and TOC) domain transcription factor (ZCCT) (Yan et al., 2004), which is induced by long days (Karsai et al., 2005; Dubcovsky et al., 2006; Trevaskis et al., 2006). Winter barleys combine a recessive allele at VRN-H1 and the presence of VRN-H2. In these genotypes, VRN-H1 is repressed by lack of cold. After a certain time under cold temperatures has passed, its expression induces the repression of VRN-H2, allowing promotion to flowering (Yan et al., 2004; Trevaskis et al., 2006; Oliver et al., 2013). On the other hand, dominant VRN-H1 alleles, carried by spring genotypes, present a constitutive expression, even without vernalization (Hemming et al., 2009). Plants with a recessive VRN-H1 and lacking VRN-H2 are described as having a facultative growth habit (Muñoz-Amatriaín et al., 2020). They present a minor vernalization response, and can be sown either in autumn or spring. VRN-H3 is the barley ortholog of the Arabidopsis FLOWERING LOCUS T gene (Yan et al., 2006). VRN-H3 integrates the flowering signals from the photoperiod and vernalization pathways (Kikuchi et al., 2009).
Two main photoperiod response genes in barley, PPD-H1 and PPD-H2 map to chromosomes 2H and 1H, respectively. PPD-H1 (HvPRR37) regulates flowering under long days (Turner et al., 2005; Campoli et al., 2012) and PPD-H2 (HvFT3) under short days (Faure et al., 2007; Kikuchi et al., 2009). The ancestral, dominant PPD-H1 allele accelerates flowering in long days (LDs), and is the most frequent in winter barleys. A natural mutation in the CCT domain of the PPD-H1 locus is associated with late flowering time under LDs (Turner et al., 2005), and is prevalent in spring barleys.
Furthermore, an interaction between high ambient temperatures and photoperiod has been reported (Hemming et al., 2012). Ejaz and von Korff (2017) showed that, at high temperatures (28°/24°C day/night), lines carrying the sensitive PPD-H1 allele, in conjunction with a spring VRN-H1 allele, accelerated plant development, whereas it was delayed in the presence of insensitive ppd-H1 and winter vrn-H1 allele. These findings suggest that PPD-H1 and VRN-H1 interact to control the development under high temperatures.
In this study, we aim to analyze the combined effects of diverse environmental conditions, namely optimum or high temperatures and presence or absence of vernalization, in a set of barley isolines with different alleles of VRN-H1, VRN-H2 and PPD-H1 on development, yield components and gene expression.
Materials and Methods
Plant Material and Growth Conditions
Eight near isogenic lines (NILs) were used in this study. These lines were developed at CSIRO Agriculture and Food (Canberra, Australia), after five rounds of crossing, up to BC4, using different donors of VRN-H1, VRN-H2 and PPD-H1 alleles into the facultative barley cultivar “WI4441” (Oliver et al., 2013). Additionally, two controls were grown, Dicktoo and Kompolti korai (Karsai et al., 2001, 2008). Dicktoo is a facultative genotype, while Kompolti korai has winter growth habit. Both of them are photoperiod-sensitive genotypes. Photoperiod-sensitive and insensitivity barleys exhibit a quantitative response to photoperiod. All barleys accelerate the development under long photoperiods, but not at the same rate. Under long days, photoperiod–sensitive genotypes accelerate the rate of development more than photoperiod-insensitive genotypes. More details about the lines are shown in Table 1 and Supplementary Figure 1.
Table 1. Genetic constitution of the barley genotypes analyzed in this study, at VRN-H1, VRN-H2, PPD-H1 and PHYC, and growth habit.
The experiment was carried out in the Phytotron facilities of the Agricultural Research Institute, Hungarian Academy of Sciences, Martonvásár, using CONVIRON PGR-15 growth chambers (Conviron Ltd., Canada). Plants were grown in pots (one plant per pot), 12 cm in diameter and 18 cm in height, with a soil capacity of 1.5 kg filled with a 4:1 mixture of garden soil and sand.
The study consisted of the factorial combination of two vernalization treatments (vernalized or unvernalized) and two temperature regimes (constant 18°C or 25°C). Seeds were planted in jiffy pots. After emergence, the plants were moved to the vernalization chamber for 60 days, at 3–4°C, under 8 h photoperiod and low light intensity [12–13 μmol m–2 s–1 photosynthetic photon flux density (PPFD)]. When the vernalization treatment was completed, plants were transplanted into individual pots and transferred to two growth chambers set at constant temperatures of either 18°C (close to the optimum level) or 25°C (above the optimum temperature level), both under 16 h of light and an intensity of 240 μmol m–2 s–1 PPFD, provided by Tungsram HGL-400 metal halide bulbs. Seeds for the non-vernalized treatments were germinated in jiffy pots, at room temperature, 7 days prior to the start of the experiment, and were transplanted to individual pots at the same time as the vernalized plants. In order to match starting points of development, a correction was performed, as all unvernalized plants were consistently slightly less developed than their counterparts in the vernalized treatments. The correction considered the difference in the initial number of leaves of the vernalized and unvernalized treatments. Through the analysis of dynamics of leaves of unvernalized treatments, we estimated the thermal time at which the number of leaves matched that of the vernalized treatment, for each genotype and temperature. These values of thermal time were subtracted from the developmental variables recorded at the unvernalized treatment. In this way, plants from both vernalization treatments started the cycle with the same number of leaves. Developmental phases Z31 (first node detectable, starting of stem elongation), Z31-49, and Z49 (awn appearance), determined according to the decimal code developed by Zadoks et al. (1974), were corrected with this method in both unvernalized treatments. Four replicates (individual plants) per genotype and treatment were used.
Measurements
Besides phenological stages Z31 and Z49, number of leaves per main stem were recorded twice per week in four plants per genotype and treatment (Haun, 1973). With these records, the dynamics of leaf appearance per genotype were determined. The number of leaves monitored in each treatment was plotted against thermal time. Sequential number of leaves for each genotype were fitted by linear regression to calculate phyllochron.
At maturity, grain yield, number of spikelets per main spike, number of grains per spikelet and per main spike, and thousand grain weight were measured in the main shoots of four plants per genotype and treatment.
Statistical Analysis
Analyses of variance (ANOVA) were performed for all the traits within each treatment using GenStat (18th Edition, VSN International, Hemel Hempstead, United Kingdom). The ANOVA models included temperature, vernalization, and genotypes, as fixed factors, and all their possible interactions, and replicates nested within temperature and vernalization treatments. The factor genotype was further divided into the three genes, VRN-H1, VRN-H2, PPD-H1, and their interactions. Several models were run, differing in the set of near isogenic lines considered in each one. Since C01 and C02 did not reach Z31 nor Z49 by the end of the experiment when not vernalized, the first model included the eight NILs, with missing values for C01 and C02 in unvernalized treatments (Table 2). The end date of the experiment was given as proxy to facilitate carrying out a statistical analysis with the full set of data (estimation for 18°C 1,800°Cd, and for 25°C 2,500°Cd, respectively). The second model also involved the eight NILs, but including the estimated values of Z31 and Z49 for unvernalized C01 and C02, Supplementary Table 1). The third model was restricted to NILs C03 to C08 (Supplementary Table 2), whilst the fourth was restricted to NILs C05 to C08 (Supplementary Table 3).
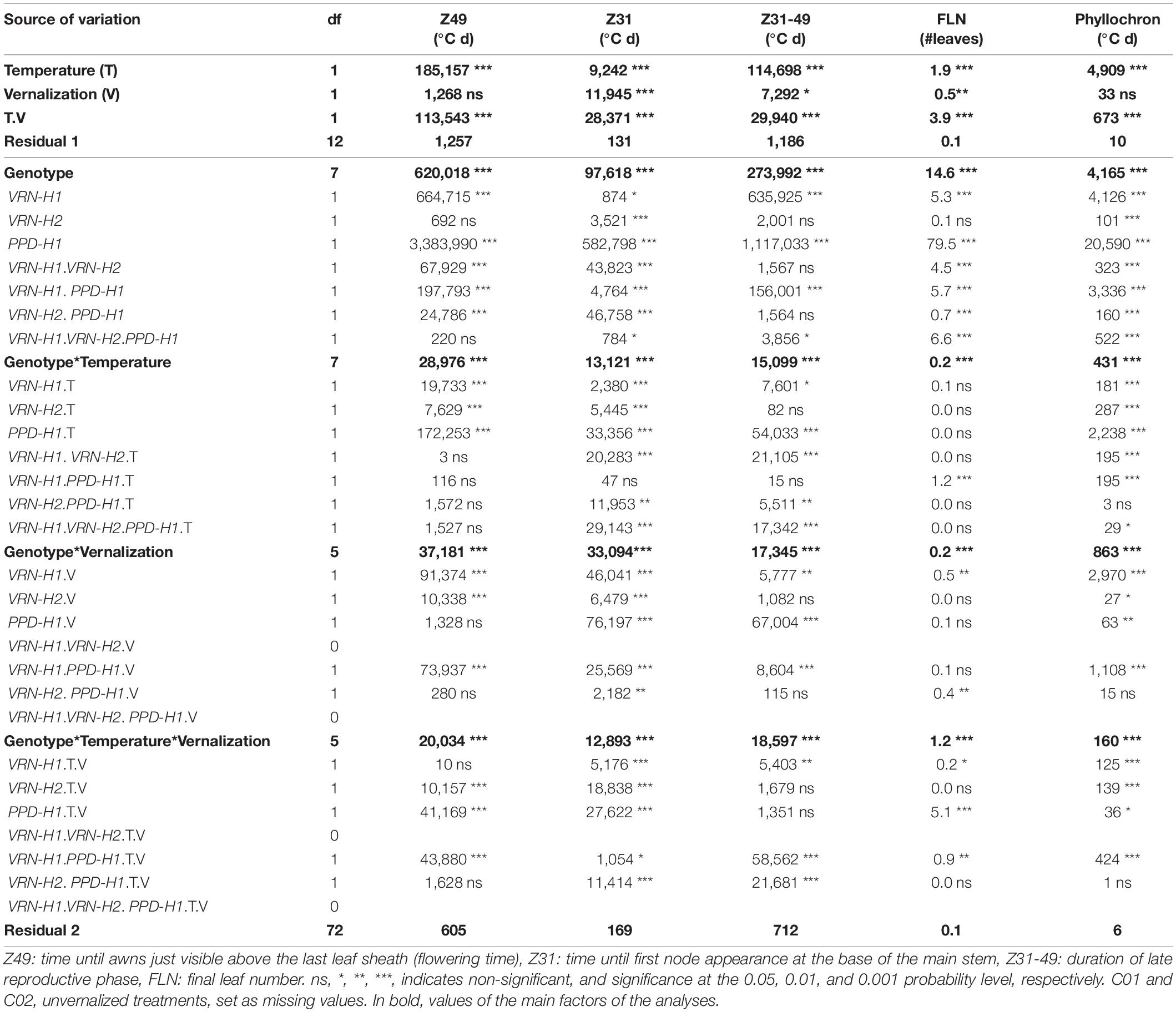
Table 2. Mean squares from the analyses of variance for the main developmental traits, considering only the eight NILs.
A correlation network analysis was carried out with the R package “qgraph” (Epskamp et al., 2012). A principal multiple factorial analysis (MFA) was performed using the R package FactoMineR (Lê et al., 2008). The R package Factoextra (Kassambara and Mundt, 2020) was employed for extracting and visualizing the results. The MFA summarizes the observations described by a set of variables structured into three groups (Phenology-development, Yield, Gene expression). These groups gather the quantitative variables measured in each experiment. Each variable within a group was equally weighted, so the influence of each set of variables in the analysis was balanced.
Genotyping
Genomic DNA was extracted from ground frozen leaves of individual plants with the EchoLUTION Plant DNA kit (BioEcho Life Sciences, GmbH). Genotyping was carried out first with diagnostic markers for the main flowering time genes, using gene specific primers for VRN-H1, VRN-H2 and PPD-H1 as reported (Turner et al., 2005; von Zitzewitz et al., 2005). Other genes analyzed were PPD-H2 (candidate gene for HvFT3), HvCEN and VRN-H3, following previously published protocols (Mansour et al., 2018). Variation in PHYC was assessed using a diagnostic KASP marker for the T/C SNP in exon 1 as described by Hill et al. (2019). All near isogenic lines used in this study have dominant alleles of PPD-H2, haplotype II of HvCEN, and the combination of late promoter, and SNPs TC (early) in the first intron of VRN-H3. Besides, whole genome genotyping of the 8 NILs was performed with the 50k Illumina SNP chip (Bayer et al., 2017), as shown in Supplementary Figure 1.
During seed multiplication, lines C01 to C04 flowered surprisingly earlier than lines C05-08, even after being vernalized (Supplementary Figure 2). Since it is known that VRN-H1 is closely linked to PHYC, a known source of earliness in barley (Nishida et al., 2013; Pankin et al., 2014), we genotyped PHYC in the NILs with a diagnostic marker. It turned out that lines C01-C04 carry the PHYC-early allele (C, from here on, PHYC-e, derived from Haruna Nijo in the WI441 genetic background, Laws et al., 2010), linked to the wild-type winter vrn-H1 allele, whereas C05-C08 have the PHYC-late allele (T, from here on referred to as PHYC-l), linked to the introgressed VRN-H1-7 spring allele. Linkage between VRN-H1 and PHYC prevents full separation of the effects of these genes in this study but, on the other hand, allows gaining insight on the performance of the latter when subjected to variable environmental conditions.
Gene Expression
For RNA extraction, leaf tissue was harvested at 0, 250 and 350°C d from the onset of temperature treatments. Samples were collected from the last fully expanded leaf, at the middle of the day (8 h after lights were turned on), immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated using Trizol Reagent (Thermo Fisher Scientific, Ltd.) and the Qiagen RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer instructions. Synthesis of cDNA was performed with 1 μg of total RNA using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific Ltd.), with the standard protocol provided by the company. qRT-PCR was carried out for three biological, and two technical replicates, in a Rotor-Gene Q equipment (Qiagen, Hilden, Germany) using the SYBR-Green technology and the corresponding standard protocol provided by the company. Expression was calculated using the Rotor-Gene software, which also considers the amplification efficiency. Relative expression was normalized against the geometric mean of two housekeeping genes, Actin and DCP5, according to Vandesompele et al. (2002). To test whether VRN-H2 expression occurred at any point during the light period, NILs C05 and C06 were grown under long photoperiod (16 h light/8 h dark) and constant temperatures of 18 and 25°C without prior vernalization, in an independent experiment. Leaf samples, three biological replicates, were sequentially collected every 3 h, starting from 3.5 to 15.5 h of light, for a total of five data points, 13 days after sowing. RNA was extracted using the Total RNA Mini Kit (IBI Scientific), and quantified using a NanoDrop 2000 (Thermo Fisher Scientific). The synthesis of first-strand cDNA was carried out with SuperScript III (Invitrogen) and derived from 1 μg RNA. Primer sequences used for qRT-PCR are showed in Supplementary Table 4.
Results
Development
Temperature, vernalization, and genotype had great impact on development. There were large and significant differences in thermal time until flowering (Z49) and its component phases (Z31 and Z31-49), due to these three factors (Figure 1, Table 2, and Supplementary Figure 3).
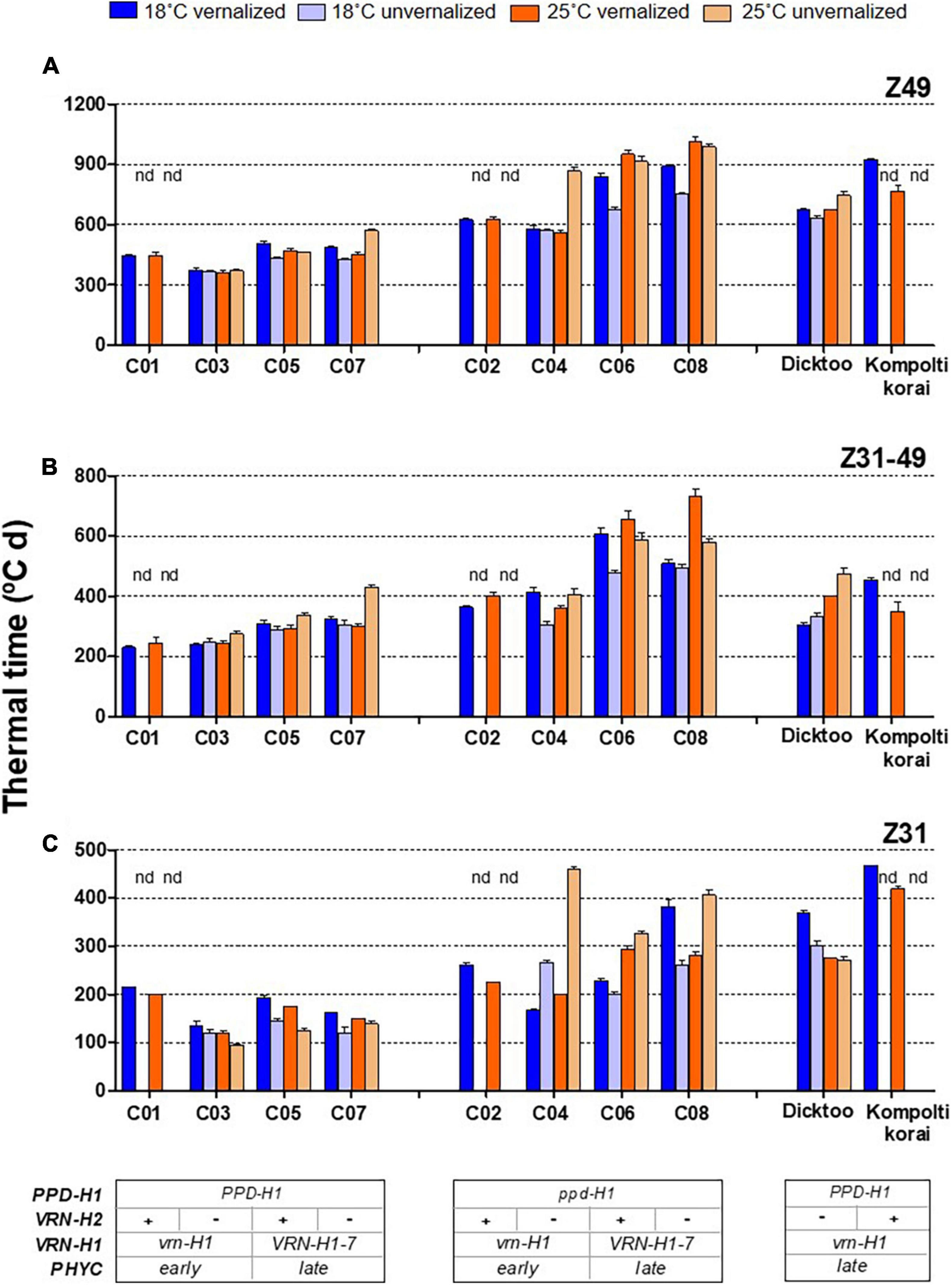
Figure 1. Plant growth duration from the onset of the experiment until flowering (A) from onset of stem elongation until flowering time (B) and from the onset of experiment until the appearance of the first node (C) for different barley NILs, whose genetic background is indicated at the bottom of the figure (inset tables). The four treatments, combinations of vernalization or absence of vernalization and temperature (18°C or 25°C) are color coded. Lines C01, C03, C05 and C07 carry dominant PPD-H1 alleles, whilst C02, C04, C06 and C08 carry a recessive ppd-H1. “nd” means not-determined. Dicktoo and Kompolti korai are check cultivars. Bars represent mean ± standard error of the mean (SE).
Regarding growth habit, isogenic lines C01 and C02 are winter types, C03 and C04 are facultative (winter allele at VRN-H1 and absence of VRN-H2), and lines C05-C08 are spring types. The patterns of development for the winter barleys essentially followed the expectations based on growth habit. Lines C01, C02, and Kompolti korai did not reach Z31 without vernalization, as their combination of winter alleles at VRN-H1 and VRN-H2 elicits a strong and compulsory vernalization response (Figure 1 and Supplementary Figure 4). We assigned C01 and C02 estimated Z31 and Z49 values of 1,800 and 2,500°C d, at 18°C and 25°C unvernalized treatments, respectively, as they correspond to the date of termination of the experiment. The results of the analyses including these estimates are presented in Supplementary Table 1. These estimates were not used in the analyses included in the main text, unless stated otherwise.
The large variation detected for flowering time, its component phases, final leaf number (FLN) and phyllochron was dominated by differences between genotypes, and to a lesser extent, though still highly significant, by temperature and interactions of genotype with both temperature and vernalization (Table 2). The only exception was phyllochron, whose variation was slightly more affected by temperature than by genotypes.
The effect of vernalization (V) was remarkable on the phase until Z31, and small afterward, with null effect on the total duration until Z49 (Table 2 and Supplementary Figure 3). At 18°C, genotypes responded differently to vernalization: time to Z49 was almost unchanged in facultative isolines (C03, C04), whereas spring lines C05-C08 and check Dicktoo suffered a slight delay due to vernalization (Figure 1A). However, at 25°C, there were few differences due to vernalization except in lines C04 and C07. These observations evidenced a significant temperature × vernalization × genotype interaction (Table 2 and Figure 1A). Overall, temperature (T) induced large variations on flowering time and its two phases (Table 2 and Figure 2). High temperature consistently increased the length of phase Z31-49, but it increased the length of the phase until Z31 only for unvernalized plants (Figure 2). The delay in flowering time caused by temperature was not consistent across vernalization and genotypes, as indicated by the large interactions found (Table 2, Supplementary Tables 1, 3, and Supplementary Figure 3). There was a significant V × T interaction, due to a crossover effect: vernalized plants had a longer cycle at 18°C and a shorter one at 25°C than unvernalized ones (Figure 2). Significant vernalization and vernalization-by-temperature effects on the time until Z31 were still detected in the analysis limited to purely spring lines C05 to C08. This is surprising, given that all the genotypes with any theoretical response to vernalization were excluded from this analysis (Supplementary Table 3).
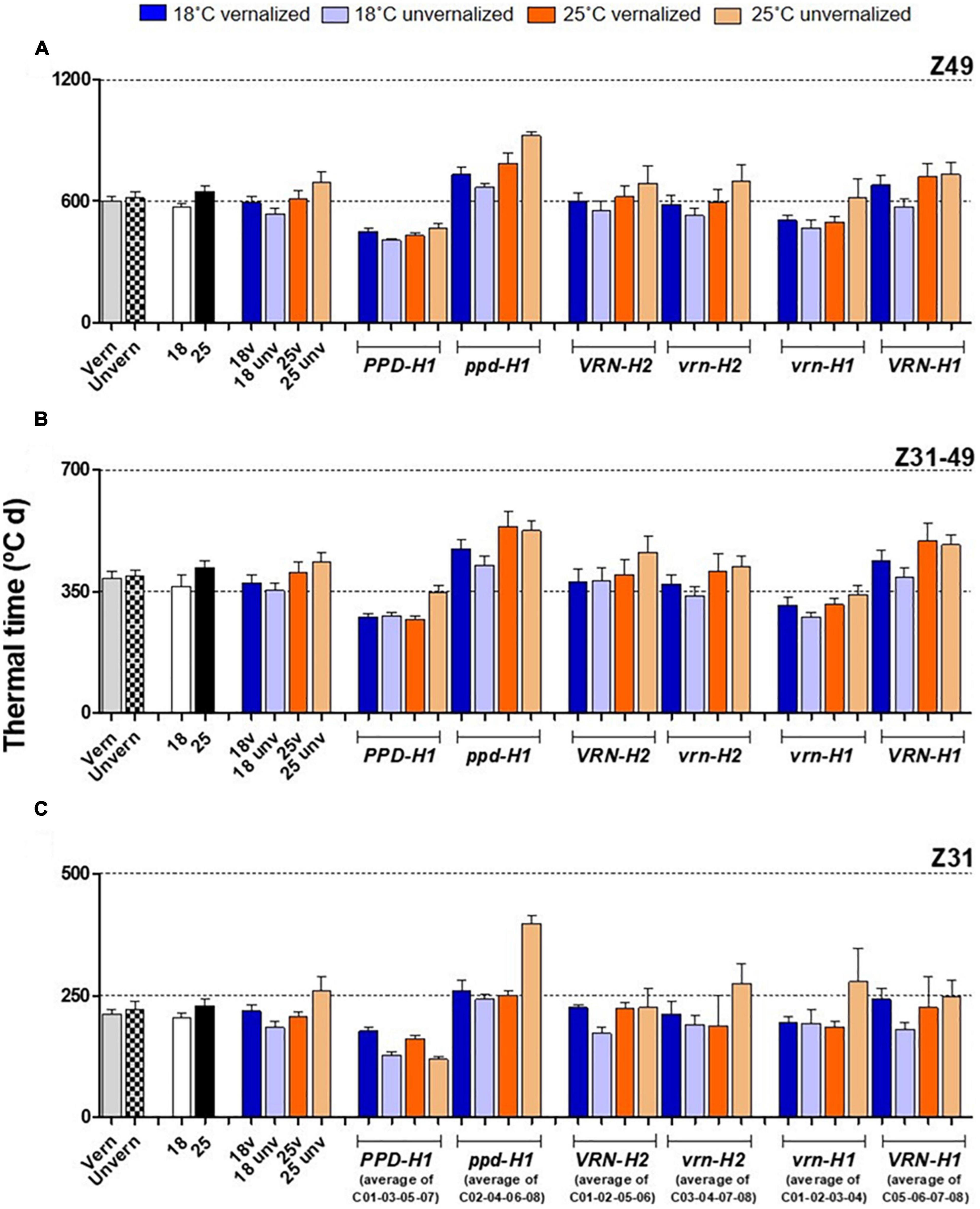
Figure 2. Duration of plant growth phases, with genotypes averaged across classes. The first 8 bars on the left of each panel represent averages of the 8 barley NILs, split by treatments: vernalized and unvernalized, 18°C and 25°C, and the four treatments combining vernalization and temperature. The other bars represent averages of the 4 NILs carrying particular alleles at the three genes polymorphic in these NILs, as indicated in the legend of the X-axes: lines classified by PPD-H1 alleles; lines classified by VRN-H2 alleles, and lines classified by VRN-H1 alleles. Duration since the onset of the experiment until flowering (A) from onset of stem elongation until flowering (B) from the onset of the experiment to the appearance of the first node (C). Bars represent mean ± standard error of the mean (SE). Means were calculated without unvernalized C01 and C02 lines.
Flowering time (Z49) was more related to the duration of the late reproductive phase (Z31-Z49), and to the phyllochron, than to changes in the duration of the phase until Z31, or final leaf number (Figure 3). Phyllochron was closely related to the duration of the late reproductive phase (r = 0.91, P < 0.001, Supplementary Figure 5A). It increased remarkably at the high temperature treatment, whereas it was roughly unchanged by the vernalization factor (Figure 4A). FLN was less affected in general by the treatments. It is worth noting that vernalization reduced FLN only at 25°C, not at 18°C, producing a significant temperature by vernalization interaction (Table 2 and Figure 4B).
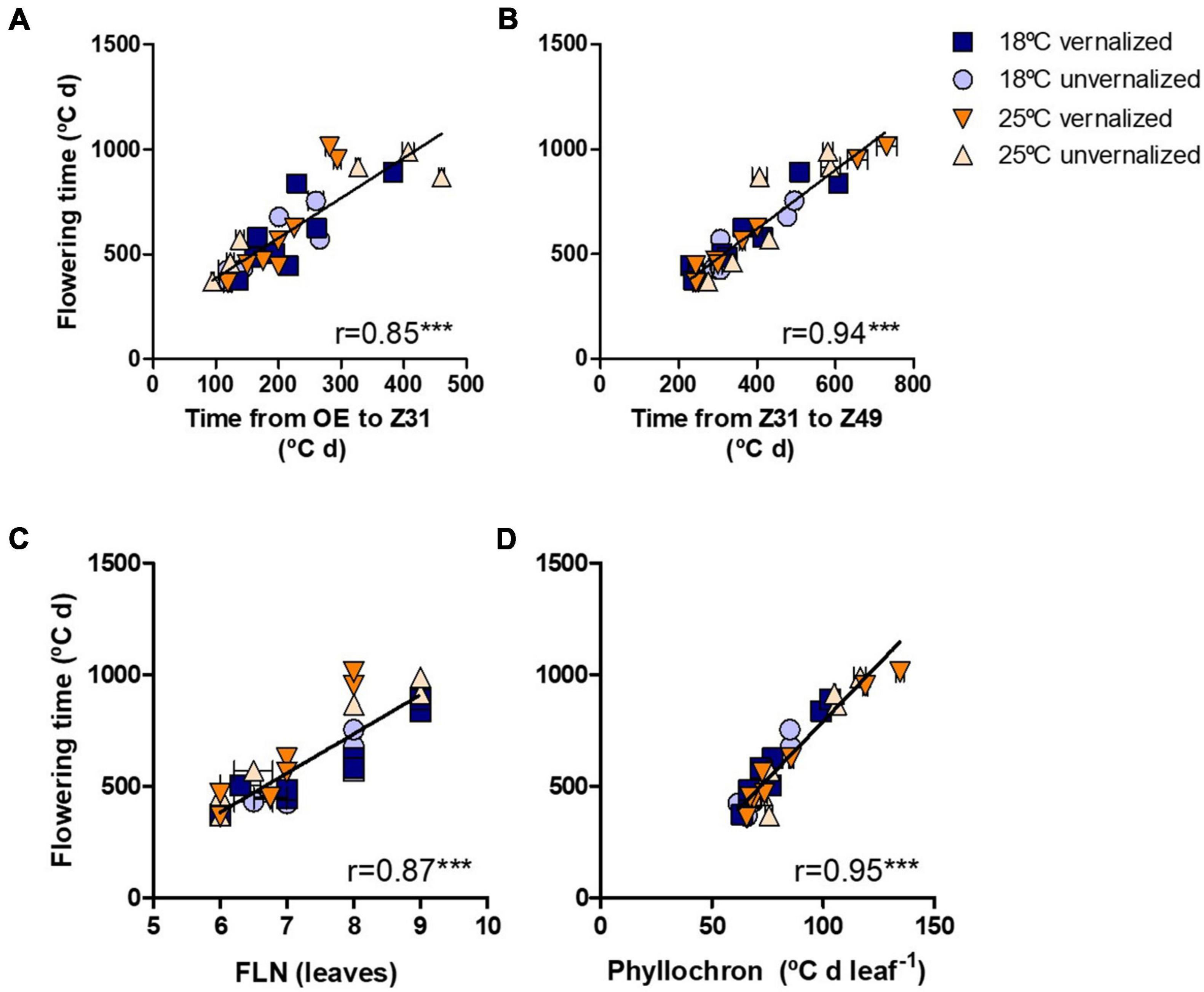
Figure 3. Relationship between time until flowering (Z49) and (A) duration from OE-onset of experiment to onset of stem elongation (Z31), (B) duration of the late reproductive phase (Z31-49), (C) final leaf number, and (D) phyllochron for eight NILs subjected to non-vernalized conditions at 18°C (circles) or 25°C (triangles) and those which were vernalized and then grown at 18°C (squares) or 25°C (down-triangles).
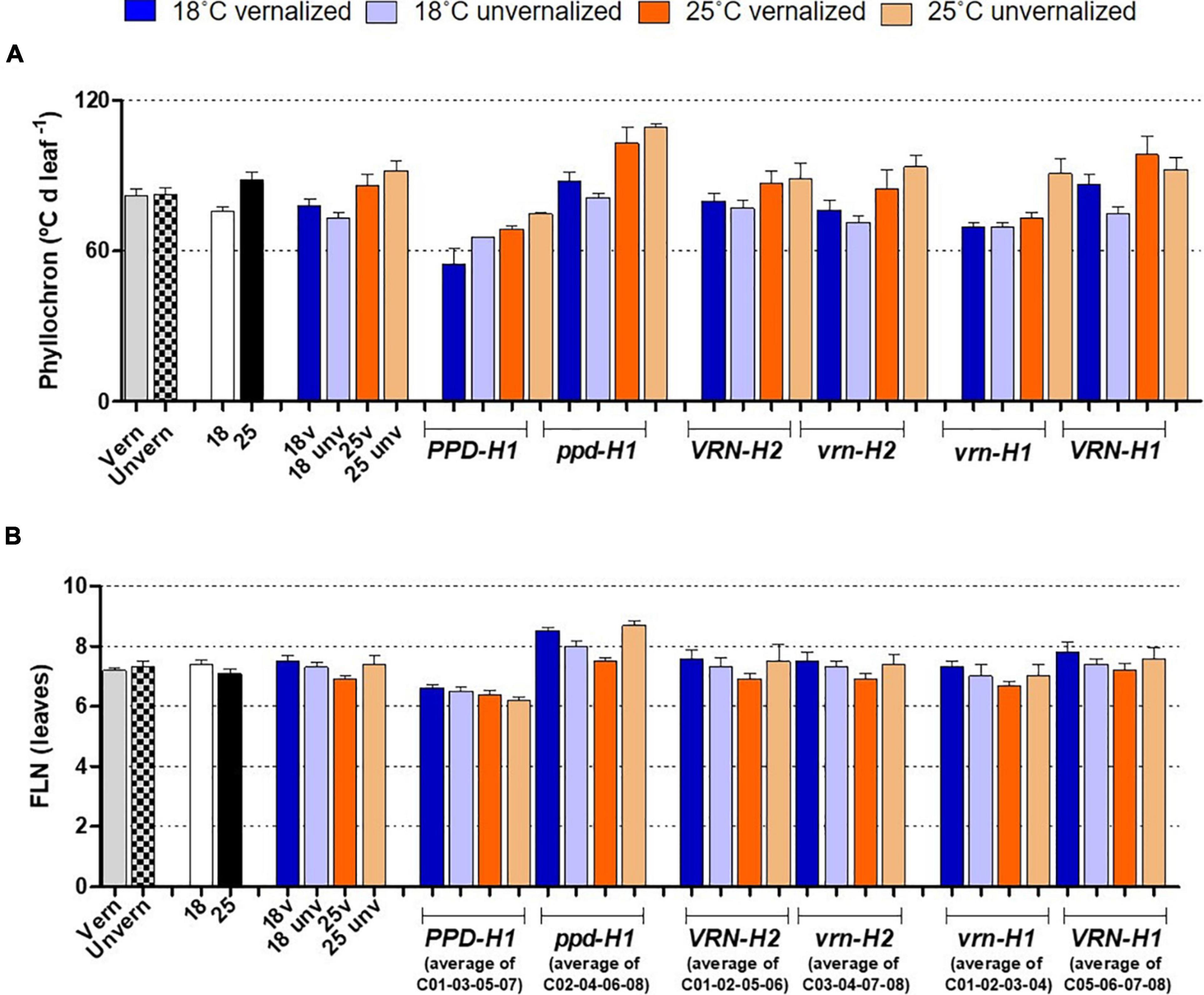
Figure 4. Phyllochron (A) and final leaf number (B), with genotypes averaged across classes. The first 8 bars on the left of each panel represent averages of the 8 barley NILs, split by treatments: vernalized and unvernalized, 18°C and 25°C, and the four treatments combining vernalization and temperature. The other bars represent averages of the 4 NILs carrying particular alleles at the three genes polymorphic in these NILs, as indicated in the legend of the X-axes: lines classified by PPD-H1 alleles; lines classified by VRN-H2 alleles, and lines classified by VRN-H1 alleles. Bars represent mean ± standard error of the mean (SE). Means were calculated without unvernalized C01 and C02 lines, which failed to reach Z31 stage.
The effect of PPD-H1 was the main driver of genotypic variation in this experiment, being the single most relevant factor in the analyses of variance for most traits (65, 55, 46, 65 and 47%, for flowering time, Z31, Z31-49, FLN and phyllochron, respectively), (Table 2, Supplementary Figure 3, and Figure 1). Plants carrying the insensitive ppd-H1 displayed delayed flowering across treatments (Figures 1A, 2A), between pairs of isolines (Supplementary Figure 6). In particular, high temperature caused the highest developmental delay of these lines when not vernalized at 25°C (255°C d), an effect 5 times larger than the one observed in the same lines when vernalized (54°C d, Figure 2).
The development of plants with sensitive PPD-H1 was almost unchanged by temperature when vernalized (452 vs. 431°C d on average at 18°C and 25°C, respectively). However, when not vernalized at 25°C, they suffered a slight but significant average delay of 59°C d until Z49, very similar to the average delay experienced by ppd-H1 recessive plants at 25°C when vernalized (54°C d, Figure 2).
The allelic effect of PPD-H1 was detected in the two stages of development, but it was larger in general on the second phase (Z31-49, Table 2 and Figure 2). However, there was a marked allelic effect in the first phase due to the delay of recessive lines when lack of vernalization was combined with high temperature (Figure 1 and Supplementary Figure 5). These fluctuations of effects underpinned the significant high order interactions of PPD-H1 with temperature, vernalization and VRN-H1 observed for Z49, Z31 and Z31-49 (Table 2).
The high temperature effect for the genotypes carrying the PPD-H1 dominant allele was small, only visible at line C07 unvernalized. For the ppd-H1 allele, the temperature effect was also very low for winter and facultative genotypes (C02 and C04) when vernalized (Figure 1). When unvernalized, line C04 suffered a remarkable delay in development, caused by a lengthened phase until Z31. However, for spring ppd-H1 lines (C06 and C08), in the unvernalized treatments, there were large temperature effects at both developmental phases, whereas when vernalization was applied, the temperature effect was, on average, large at the Z31-49 phase, and almost negligible for Z31. In this last case, however, the effect was the average of two very different responses for the period until Z31, of the two spring lines carrying the recessive ppd-H1 allele (C06 and C08). The reaction of PPD-H1 alleles to temperature was complex and dependent on other factors. A general conclusion is that the dominant allele of PPD-H1 and vernalization acted additively to buffer the effects of an increasing temperature on development.
The effect of recessive ppd-H1 on development was a combination of larger FLN and lengthening of the phyllochron (Figure 4 and Supplementary Figure 6), modified by temperature and vernalization, and specific genotypic effects.
There was no overall effect of VRN-H2 on Z49, but it did affect the duration until Z31 and presented significant interactions with temperature and vernalization (Table 2). In spring lines (C05-C08), the effect of VRN-H2 depended on the PPD-H1 allele present.
In spring lines, C05-C08, the VRN-H2 × PPD-H1 interaction affected total duration (Z49), but through different phases (Figure 1 and Supplementary Table 3). In general, presence of VRN-H2 shortened the first phase (Z31) in recessive ppd-H1 lines, and the second one (Z31-49) in PPD-H1 dominant lines (mostly due to the 25°C unvernalized treatment). VRN-H2-carrying lines had shorter cycle than VRN-H2-absent lines (41°C d) overall, but particularly in presence of recessive ppd-H1 (67°C d vs. only 16°C d in presence of dominant PPD-H1). However, this effect was mostly overridden at 25°C, and was independent of vernalization.
Regarding VRN-H1, leaving out unvernalized C01 and C02, plants with the winter vrn-H1 allele flowered earlier than those carrying spring VRN-H1-7 (Figure 2). This result was partly due to the presence of the PHYC-e allele, completely linked to the winter vrn-H1 allele. Yet, plants C01 and C02 did not flower without vernalization, indicating that the effect of PHYC did not override their vernalization requirement.
The effect of PHYC on flowering can be indirectly estimated attending to the results of vernalized treatments. When vernalized, earliness due to the presence of the different VRN-H1/VRN-H2 haplotypes should disappear. Any differences remaining should be the result of other genes, most likely PHYC. There were significant differences between lines split by the VRN-H1 alleles (likely, PHYC effect) in time to reach Z49 in both vernalized treatments. At both temperatures, the effect was cumulative over the two developmental phases, but was more marked on the second one (Z31-Z49) (Figure 2).
Inflorescence Traits
Inflorescence traits recorded in each main spike were grain yield, grain number, number of spikelets, thousand grain weight, and grain number per spikelet. All these traits were affected by temperature and vernalization (Supplementary Table 5). The lack of spikes in unvernalized lines C01 and C02 caused that vernalization, VRN-H1 and of VRN-H2 and their interactions were highly significant, and are the dominant sources of variation in the analysis of variance with all 8 isogenic lines, including estimates for C01 and C02 unvernalized (Supplementary Table 5). The following comments pertain to the analyses in which these large effects were removed by setting all C01 and C02 unvernalized yield and yield component scores as missing values (Table 3). The findings of this first analysis were later confirmed in analyses carried out with lines C05-C08 (all spring lines, without VRN-H1 nor PHYC polymorphism, Supplementary Table 3), and with lines C03-C08 (Supplementary Table 2).
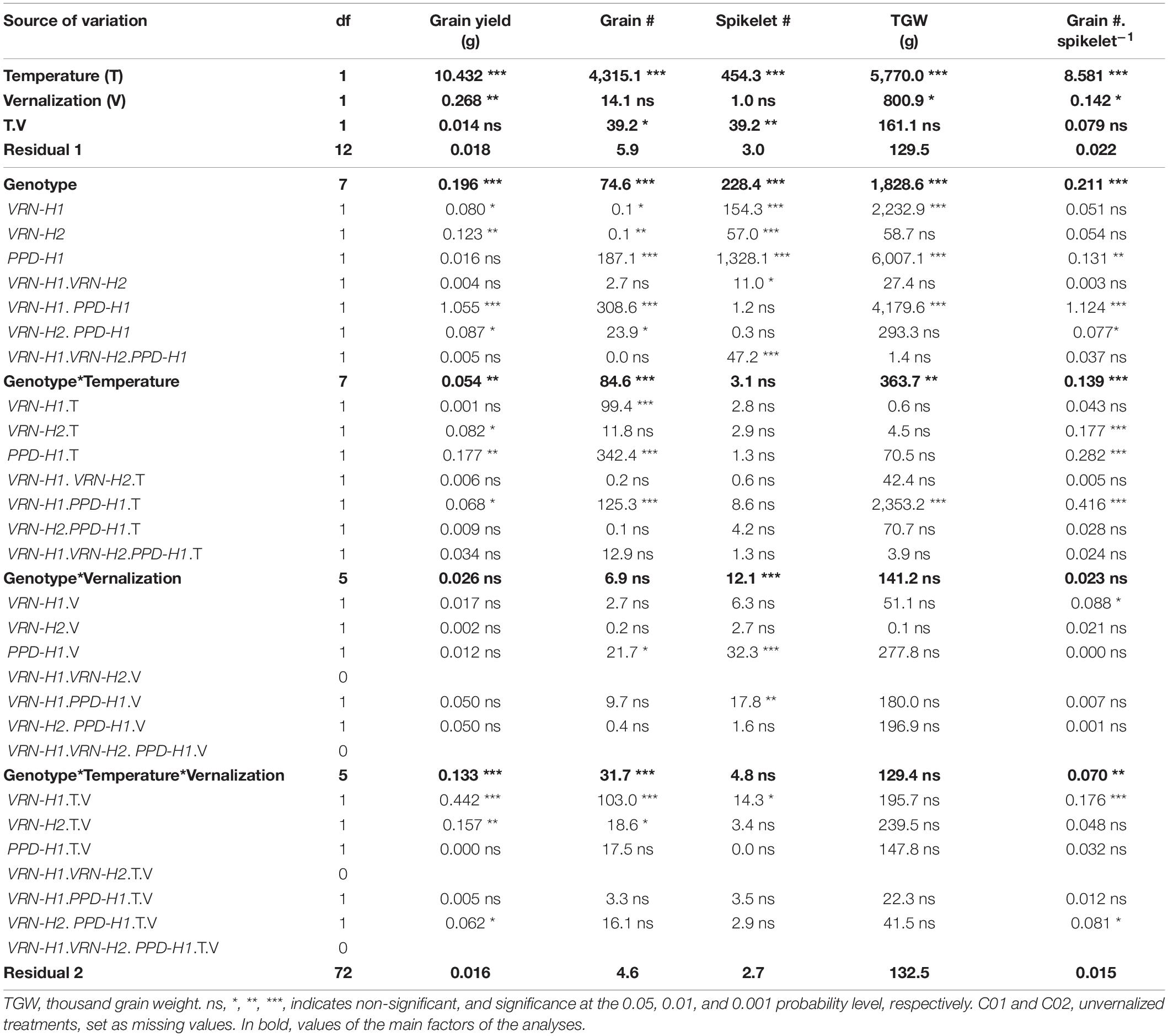
Table 3. Mean squares from the analyses of variance for inflorescence traits measured at the main spike of each plant, corresponding to the eight NILs.
Some VRN-H1 and VRN-H2 effects on yield and components were significant, particularly the main effects on grain yield and spikelets per spike but, as for the developmental traits, the most important effects on these traits were also caused by PPD-H1, except for grain yield per main spike (Table 3). Recessive ppd-H1 lines always had more spikelets per spike than their respective PPD-H1 counterparts, while the trend for TGW was opposite (higher TGW for dominant PPD-H1 lines) across all treatments (Figure 5).
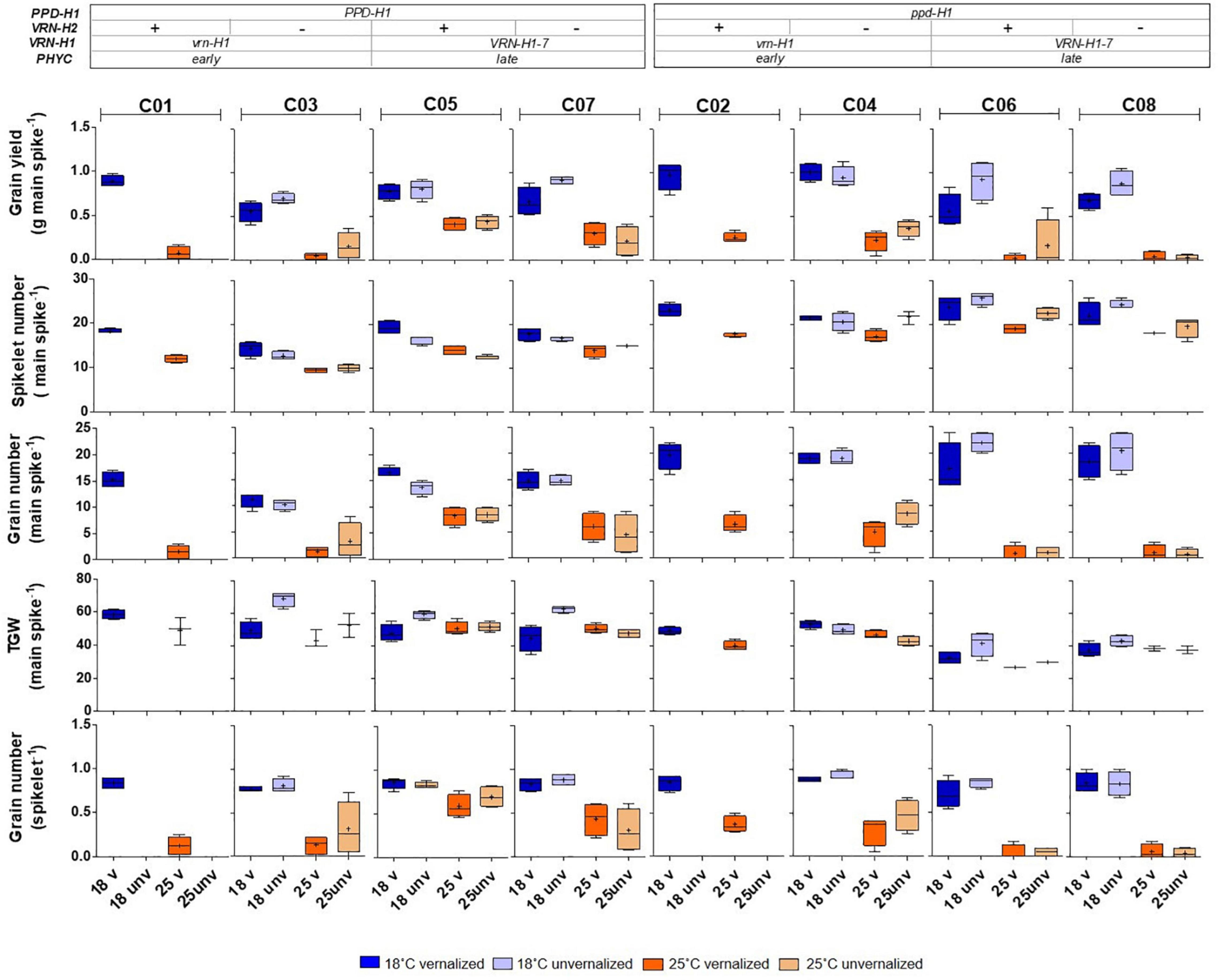
Figure 5. Inflorescence traits corresponding to the main spike: grain yield, spikelet number, grain number, thousand grain weight (TGW), and grain number per spikelet of eight barley NILs subjected to non-vernalized and vernalized conditions and grown at 18 and 25°C. The genetic composition of the lines is described in the top inset.
In addition, the interactions between PPD-H1 and VRN-H1/PHYC and PPD-H1 with temperature (interactions VRN-H1 × PPD-H1, PPD-H1 × T, VRN-H1 × PPD-H1 × T), were significant for grain yield or several yield components (Table 3). The effect of interactions between PPD-H1 and temperature and PPD-H1 and VRN-H1 (when testable) were confirmed in partial analyses involving only lines C03-C08, and lines C05-C08, so these effects were independent of growth habit. The most striking result was the large interaction between VRN-H1 and PPD-H1 for yield of the main spike (Table 3). Recessive ppd-H1 lines had higher yield in vrn-H1/PHYC-e background across all treatments, and dominant PPD-H1 lines yielded best in VRN-H1-7/PHYC-l lines overall. However, this last observation was caused by the clear yield advantage of dominant PPD-H1 lines at 25°C, whereas at 18°C there was no advantage of either allele. All lines showed a striking yield reduction at 25°C, but the largest reductions occurred in haplotypes vrn-H1/PPD-H1 and VRN-H1-7/ppd-H1. They were caused by poorer seed set and, to a lesser extent, by a reduced TGW only in VRN-H1-7/ppd-H1 lines (Figure 5).
The phenological traits of these isolines at 18°C are good predictors of their development at 25°C (correlation coefficients of 0.96 and 0.99 for Z49 in unvernalized and vernalized plants, respectively), but the predictability of yield at high temperature, based on the 18°C performance was poor (correlations of 0.44 for vernalized plants, and 0.08 for unvernalized ones). For yield components, the number of spikelets per main spike and TGW present the largest correlation coefficients between temperatures (0.93 and 0.68, respectively), whereas the correlations for grain number per spike and grain yield per main spike were close to 0. The duration of the phase until the appearance of the first node (Z31), determines the number of spikelets. The lines with the insensitive ppd-H1 allele presented a delayed appearance of the first node and, fittingly, they are characterized by a higher spikelet number per spike than lines with the sensitive PPD-H1 alleles. However, insensitive ppd-H1 lines also presented lower grain number in the background of spring VRN-H1-7 alleles at high temperature, indicating a reduced spike fertility.
Gene Expression
As expected, winter barleys (C01 and C02) exhibited VRN-H2 expression mostly in unvernalized plants, in absence of its VRN-H1 repressor (Figure 6). Initially, VRN-H2 transcripts were not detected in spring barleys in this experiment. However, we had previous data indicating that this gene was expressed in spring lines C05 and C06 (Monteagudo, personal communication). We grew these lines under two temperatures and in absence of vernalization in a separate experiment. We found a small but clear VRN-H2 expression, apparently following a circadian rhythm, with maximum expression at the end of the day period, higher in C05 than in C06 (Supplementary Figure 8). Therefore, a phenotypic effect of VRN-H2 expression in these spring lines cannot be ruled out.
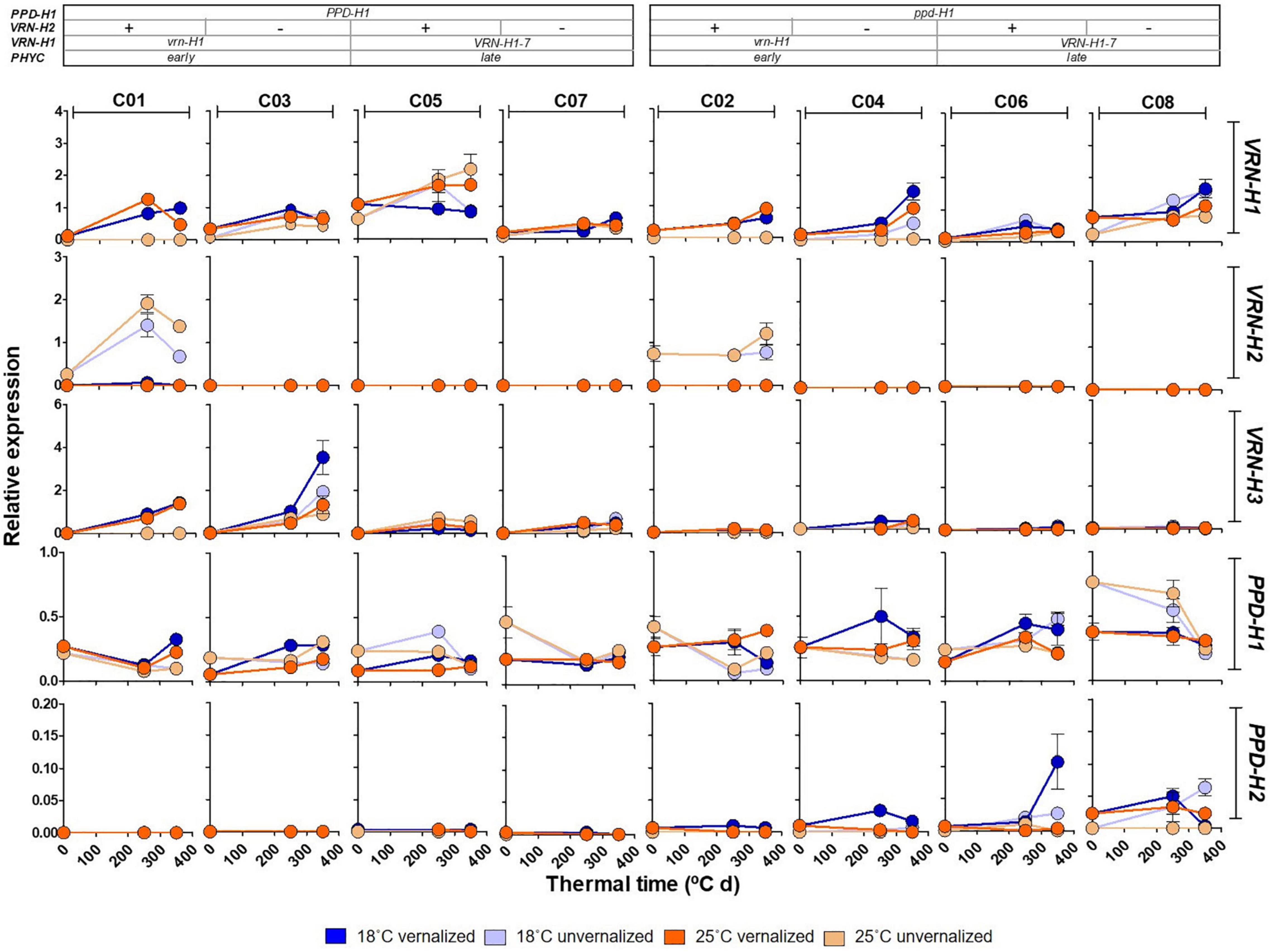
Figure 6. Expression patterns of VRN-H1, VRN-H2, VRN-H3, PPD-H1 and PPD-H2 at three different times (0, 250 and 350°C d), for the eight barley NILs (C01-C08) subjected to non-vernalized and vernalized conditions and grown at 18 and 25°C. The genetic composition of the lines is described in the top inset.
All the lines had the same VRN-H3 and PPD-H2 alleles. However, their expression was strongly dependent on the PPD-H1 allele present, with lines carrying the sensitive allele showing higher VRN-H3 expression. Expression of PPD-H2, the “short-day sensitivity” gene was detected, although the experiment was carried under long days. Contrary to VRN-H3, PPD-H2 expression was higher in recessive ppd-H1 lines (Figure 6). PPD-H2 expression also varied widely with the experimental factors, with highest expression in vernalized plants at 18°C.
Patterns of PPD-H1 expression were highly variable across genotypes (Figure 6). Overall, the insensitive allele of PPD-H1 showed higher expression than the sensitive one (Figure 6 and Supplementary Table 6). These results may be affected by the specific sampling time, as the circadian rhythm of the two PPD-H1 alleles may reach expression peaks at different times (Campoli et al., 2012), as also hinted in our result with sequential samples at 18°C (Supplementary Figure 8). PPD-H1 expression is modulated by the allele present at VRN-H1/PHYC, as indicated by a significant interaction (Supplementary Table 6).
Gene expression of flowering promoters VRN-H1 and VRN-H3 was negatively correlated with development variables (Supplementary Figure 5), whereas the opposite was true for that of PPD-H1 and PPD-H2 (which showed high interdependence, as mentioned before). Yield and grain number, on the other hand were rather independent of development and gene expression variables. Late lines showed an increased number of spikelets per spike, but lower TGW (Figure 5).
Discussion
This set of isogenic lines constitutes an excellent genetic screen to test the joint effects of developmental genes. The components of the vernalization and photoperiod pathways in barley are rich in interactions and, therefore, studying their effects in isolation may not reveal their roles in full. In fact, this study has unraveled a complex grid of interactions between the main genes of the vernalization and photoperiod pathways, and the environmental conditions, some of them unexpected.
Which Gene Has the Largest Effect on Development?
The performance of winter lines C01 and C02, was in agreement with the epistatic mode of action of the vernalization mechanism in winter varieties proposed by Trevaskis et al. (2006). Aside from the failure of winter lines to progress toward flowering without vernalization, PPD-H1 had the largest effect on development, irrespective of temperature. This finding is in accordance with other studies carried out with barley (Turner et al., 2005; Digel et al., 2016; Wiegmann et al., 2019; Gol et al., 2021).
We could not separate the effects of VRN-H1 and PHYC fully. We could deduce, however, that the strong earliness effect induced by PHYC-e (Nishida et al., 2013; Pankin et al., 2014) was not able to override the need for vernalization of winter lines C01 and C02, as they did not progress toward reproductive development when not vernalized. The protein-protein interaction between PHYC and VRN2 described in wheat by Shaw et al. (2020) may have played a role, as VRN-H2 was expressed in abundance only in those genotypes and condition.
We have observed a larger role of vernalization on the initial phase of the cycle, until onset of stem elongation (Z31), than on the late reproductive phase (Z31-49). Yet, vernalization effects were still appreciable in the second phase, as previously reported by González et al. (2002). In contrast, temperature affected the duration of the entire cycle, but more markedly the late reproductive phase, in accordance with previous findings in wheat (Slafer and Rawson, 1994). However, Karsai et al. (2013) observed temperature effects on barley development at all growth stages, particularly around the appearance of the first node.
Apparently, vernalization acted as a stabilizing factor for the development of the lines, having more or less effect on development depending on the genetic background, and counteracted the effect of high temperature on both phenology and yield. Vernalized lines had a more stable behavior across temperatures, whereas unvernalized plants suffered developmental delays at high temperature during the two stages considered. In every case, delays in development were related with longer phyllochrons. This observation may be relevant to barley grown in autumn sowings. Cold periods are long enough to provide at least some vernalization in most areas where it is practiced.
Our high temperature treatment, 25°C is likely above the optimum temperature for barley growth. The effect of this treatment varied among genotypes and growth phases, which may be related to the reported differences in optimum temperatures for these phases in wheat and barley (Jacott and Boden, 2020). Optimum temperatures up to Z31 are lower than after that stage, and the larger effect at this stage observed for some lines could be related to a wider distance between the optimum temperature and our high temperature treatment. We found that high temperatures delayed flowering time in unvernalized plants, except line C03. It has been reported that high temperatures delay development of barley genotypes carrying the recessive ppd-H1 allele (Ejaz and von Korff, 2017), which agrees with our own results. However, when vernalized, the delay in development of ppd-H1 plants occurred only in spring barleys (C06 and C08, compared to C05 and C07, respectively). Other stress conditions, like drought, also affect spring barleys carrying insensitive ppd-H1 alleles, by delaying flowering time (Gol et al., 2021). It is sensible to speculate whether this allele presents intrinsic disadvantages under abiotic stress conditions.
We have demonstrated that the previously reported effect of temperature on PPD-H1 alleles is not constant. It depends, to a large extent, on interactions between environmental conditions (presence/absence of vernalization and ambient temperature), and the haplotype at the vernalization genes.
Traditionally, spring cultivars do not present any of the winter alleles at VRN-H1 and VRN-H2 (in a few cases, they carry just one of the two). These results indicate that a judicious use of allelic diversity at the vernalization genes, in combination with the allele at PPD-H1, would provide ample options to modulate plant development, and fine tuning it according to the expected temperatures and sowing times. Different haplotypes will either induce a longer vegetative phase, or a longer phase after onset of stem elongation. Which strategy is best to enhance grain yield production at each environment, will have to be ascertained with further research involving modeling for future conditions. Then, plant breeders will make use of this toolbox of alleles and known interactions with environment to construct ideotype cultivars specific to each agroecological region.
The Presence of an Active VRN-H2 Affects the Development of Spring Lines
One of the novel results of this work was the detection of an effect of VRN-H2 on development, partly through the interaction VRN-H2 × PPD-H1, which was particularly striking in the case of spring lines (C05-C08). According to the current consensus, the spring VRN-H1 allele they carry should be constitutively expressed, without the need of external cues, and should repress the expression of VRN-H2. Initially, we did not detect clear signs of expression of VRN-H2 in these lines (Figure 6). However, those samples represent a single time of the day, and we could have missed the moment of peak expression. We did a new experiment growing unvernalized lines C05 and C06, at 16 h and 18°C or 25°C, and checked gene expression (for VRN-H2 and other genes) at several time points of the day in 13-day old plants. The results, presented in Supplementary Figure 8, indicated that there was feeble but unequivocal VRN-H2 expression in both lines, more at 18°C than at 25°C. This demonstrates that VRN-H2 is expressed in spring lines under long days, and could be responsible for the effects observed. The relationship of the VRN-H2 by PPD-H1 interaction with the duration of growth until Z31 could be related with the existence of regulatory feedback loops between VRN-H2, PPD-H1 and HvCO2, described by Mulki and von Korff (2016).
The development of line C06, ppd-H1/VRN-H2 is particularly interesting, as it represents an alternative format to the typical pattern of spring cultivars (line C08). C06 ideotype may have an agronomic merit. It is earlier than C08, but has a similar duration of the phase after the onset of stem elongation as C08, which is crucial for the determination of the potential number of grains in the spike. Its earliness is due to a shorter phase until Z31. This could be a way of shortening the cycle of spring cultivars without compromising yield potential, although its yielding ability is heavily impaired at high temperature.
Growth Cycle of Spring (ppd-H1 Recessive) Lines Is Modified by Vernalization Through Changes in Phyllochron
Vernalization, optimal temperature, and the presence of an ancestral PPD-H1 allele, are all stabilizing factors for barley development. Alterations in any of them cause a wide variety of responses.
One of the unexpected results of this study was finding a PPD-H1 × V interaction for the duration of period Z31-49, in all analyses of variance performed. It is remarkable that it was one of the most relevant sources of variance in the ANOVA restricted to the spring lines (C05-C08). The spring lines with the recessive ppd-H1 allele were significantly earlier in reaching flowering when not subjected to vernalization than when vernalized (89°C d). The difference was entirely due to a longer late reproductive period of ppd-H1 recessive lines when vernalized, related to a longer phyllochron (16% longer than without vernalization). We cannot discard that there are other genes acting in the vernalization pathway still present in the genome of these isolines, interacting with vernalization and PPD-H1, to produce the results observed.
PPD-H1 and VRN-H1 Interaction Affects Grain Yield
Estimation of grain yield and its components in plants grown in growth chambers cannot be extrapolated to field conditions. Nonetheless, we expect that the trends observed could be indicative of what can also be expected in the field. We confirmed a drastic reduction of grain yield, spikelet number and grain number, in the eight lines, as a consequence of exposure to a constant high temperature throughout the cycle. In fact, short or prolonged periods of high temperatures are known to reduce barley yield (Hemming et al., 2012; Jacott and Boden, 2020), which has been confirmed in field experiments applying heat stress, both on wheat (Wollenweber et al., 2003; García et al., 2015; Elía et al., 2018), and barley (García et al., 2016), considerably reduced grain number and grain weight.
Lines carrying the sensitive PPD-H1 allele presented lower spikelet number than lines with insensitive ppd-H1 alleles, irrespective of temperature and vernalization treatments, thus showing a lower yield potential. At 18°C, plants with ppd-H1 alleles generated more grains per spike and yielded more. At 25°C, however, spike fertility and grain filling of ppd-H1 plants collapsed. The number of grains produced per spike dropped dramatically, and the few grains produced were lighter, compared to PPD-H1 carrying plants. Ejaz and von Korff (2017), also found that high ambient temperatures caused a larger reduction of seeds per spike in lines carrying insensitive ppd-H1 alleles, in comparison with those carrying sensitive PPD-H1 alleles. However, in our case, the effect was only evident in lines carrying also the spring VRN-H1-7 allele.
The consequences of these phenomena cannot be extrapolated directly to field-grown barley. However, there is sound evidence on the large impact of PPD-H1 alleles on grain yield depending on environmental conditions. Wiegmann et al. (2019) found that, where earliness was beneficial, the sensitive PPD-H1 allele provided a yield advantage. Gol et al. (2021) reported a larger yield stability under drought for plants with the sensitive allele. The study of yield impacts of this gene and its orthologs in relation with temperature should be a priority in cereal research.
PPD-H1 Alleles Associated to Transcript Levels of VRN-H3 and PPD-H2
The interactions between VRN-H2 and PPD-H1, and of these two genes with VRN-H1 had a large influence on VRN-H1 expression. The highest VRN-H1 expression in line C05 seemed to underpin these interactions (Supplementary Table 6). There is no clear explanation for this strikingly high level of VRN-H1 expression in this line, across all four treatments. Oliver et al. (2013) reported a higher expression potential for VRN-H1-7, compared to other alleles but, in our results, this was only true for line C05 and not for the other three lines with this allele. Therefore, we hypothesize that the higher expression of this VRN-H1 allele is not constitutive, and that it depends on the genetic background. Usually, this higher expression should result in upregulation of VRN-H3 (Deng et al., 2015), but this is not clearly observed in C05. This line carries, besides a spring allele at VRN-H1, a winter VRN-H2 allele and a sensitive PPD-H1 allele. According to Mulki and von Korff (2016), a sensitive PPD-H1 allele up-regulates VRN-H2 expression. Additionally, we have detected expression of VRN-H2 in C05 (Supplementary Figure 8). Therefore, we could speculate on the presence of a feedback loop mechanism inducing more VRN-H1 expression in C05 to down-regulate the VRN-H2 present in this line.
PPD-H2 was expressed in some lines, although at low levels, despite the long photoperiod. However, it is controlled by day length only indirectly, with other genes acting as intermediary (Casao et al., 2011). Actually, observing its expression in long days, as in our experiment, is not new (Kikuchi et al., 2009; Casao et al., 2011; Monteagudo et al., 2019, 2020). The clear dependence of its expression on the PPD-H1 allele observed here, was already pointed out by Mulki et al. (2018).
As expected, lines with sensitive PPD-H1 alleles showed higher transcript levels of VRN-H3 and flowered earlier compared with recessive ppd-H1 alleles (Turner et al., 2005).
The main genetic feature of spring cultivars from Central Europe is the presence of the insensitive ppd-H1 allele (Jones et al., 2011; Russell et al., 2016; Bustos-Korts et al., 2019). The appearance of the insensitive ppd-H1 allele allowed the expansion of the crop to areas in which late winter or spring sowings were required to escape the harsh winter conditions (Jones et al., 2008; Wiegmann et al., 2019). Most spring cultivars, however, also carry the active PPD-H2 allele (early), and haplotype III at gene HvCEN (late). This combination of alleles may have an adaptive purpose, which has not been explained yet. The presence of the active PPD-H2 allele, the “short photoperiod sensitivity” gene seemed irrelevant in spring sowings, because these crops complete their cycles under long days. The fact that PPD-H2, a known promoter of spikelet initiation (Mulki et al., 2018) is expressed in long days, and at higher levels in the presence of the insensitive ppd-H1 allele, suggests an active role of PPD-H2 in promotion of development even in spring sowings.
In conclusion, our study provides useful tools to better understand the physiological and molecular responses of barley at high ambient temperatures and contrasting vernalizing treatments, through several allelic combinations of VRN-H1, VRN-H2 and PPD-H1 genes. New contributions shown here might be important for breeders to develop barley varieties with improved resilience under future climate conditions.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: DIGITAL.CSIC, at http://hdl.handle.net/10261/256755. Further inquiries can be directed to the corresponding author/s.
Author Contributions
IK, AC, and EI: conceptualization, methodology, and writing—review and editing. TK, IK, and HO: investigation. HO, AC, and EI: formal analysis. AC and EI: supervision and funding acquisition. HO: visualization. HO and EI: writing—original draft preparation. All authors read and approved the final manuscript.
Funding
Funding of this research was provided was provided by project AGL2016-80967-R (Agencia Estatal de Investigación, Ministry of Economy and Competitiveness), project PID2019-111621RB-I00 (Agencia Estatal de Investigación), grants A08_17R and A08_20R (Government of Aragón), and project GINOP-2.3.2-15-2016-00029 (Economic Development Programs of the Hungarian Ministry of Finance). HO was supported by a Juan de la Cierva-Formación fellowship, granted by the Spanish Ministry of Science and Innovation (FJC2018-037885-I).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Isogenic lines C01-C08 were developed by Sarah Fieg, working at the group of Ben Trevaskis, Black Mountain Laboratory, CSIRO Canberra, Australia, who kindly provided the seed for this work. We would like to acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.776982/full#supplementary-material
References
Asseng, S., Ewert, F., Martre, P., Rotter, R. P., Lobell, D. B., Cammarano, D., et al. (2015). Rising temperatures reduce global wheat production. Nat. Clim. Chang. 5, 143–147. doi: 10.1038/nclimate2470
Bayer, M. M., Rapazote-Flores, P., Ganal, M., Hedley, P. E., Macaulay, M., Plieske, J., et al. (2017). Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 8:1792. doi: 10.3389/fpls.2017.01792
Bustos-Korts, D., Dawson, I. K., Russell, J., Tondelli, A., Guerra, D., Ferrandi, C., et al. (2019). Exome sequences and multi-environment field trials elucidate the genetic basis of adaptation in barley. Plant J. 99, 1172–1191. doi: 10.1111/tpj.14414
Campoli, C., Shtaya, M., Davis, S. J., and von Korff, M. (2012). Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12:97. doi: 10.1186/1471-2229-12-97
Cantalapiedra, C. P., García-Pereira, M. J., Gracia, M. P., Igartua, E., Casas, A. M., and Contreras-Moreira, B. (2017). Large differences in gene expression responses to drought and heat stress between elite barley cultivar scarlett and a Spanish landrace. Front. Plant Sci. 8:647. doi: 10.3389/fpls.2017.00647
Casao, M. C., Igartua, E., Karsai, I., Lasa, J. M., Gracia, M. P., and Casas, A. M. (2011). Expression analysis of vernalization and day-length response genes in barley (Hordeum vulgare L.) indicates that VRNH2 is a repressor of PPDH2 (HvFT3) under long days. J. Exp. Bot. 62, 1939–1949. doi: 10.1093/jxb/erq382
Danyluk, J., Kane, N. A., Breton, G., Limin, A. E., Fowler, D. B., and Sarhan, F. (2003). TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132, 1849–1860. doi: 10.1104/pp.103.023523
Deng, W., Casao, M. C., Wang, P., Sato, K., Hayes, P. M., Finnegan, E. J., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6:5882. doi: 10.1038/ncomms6882
Digel, B., Tavakol, E., Verderio, G., Tondelli, A., Xu, X., Cattivelli, L., et al. (2016). Photoperiod-H1 (Ppd-H1) controls leaf size. Plant Physiol. 172, 405–415. doi: 10.1104/pp.16.00977
Dubcovsky, J., Loukoianov, A., Fu, D., Valarik, M., Sanchez, A., and Yan, L. (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60, 469–480. doi: 10.1007/s11103-005-4814-2
Elía, M., Slafer, G. A., and Savin, R. (2018). Yield and grain weight responses to post-anthesis increases in maximum temperature under field grown wheat as modified by nitrogen supply. Field Crop. Res. 221, 228–237. doi: 10.1016/j.fcr.2018.02.030
Epskamp, S., Cramer, A. O. J., Waldorp, L. J., Schmittmann, V. D., and Borsboom, D. (2012). Qgraph: network visualizations of relationships in psychometric data. J. Stat. Softw. 48, 1–18. doi: 10.18637/jss.v048.i04
Faure, S., Higgins, J., Turner, A., and Laurie, D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
García, G. A., Dreccer, M. F., Miralles, D. J., and Serrago, R. A. (2015). High night temperatures during grain number determination reduce wheat and barley grain yield: a field study. Glob. Chang. Biol. 21, 4153–4164. doi: 10.1111/gcb.13009
García, G. A., Serrago, R. A., Dreccer, M. F., and Miralles, D. J. (2016). Post-anthesis warm nights reduce grain weight in field-grown wheat and barley. Field Crop. Res. 195, 50–59. doi: 10.1016/j.fcr.2016.06.002
Giménez, V. D., Miralles, D. J., García, G. A., and Serrago, R. A. (2021). Can crop management reduce the negative effects of warm nights on wheat yield? Field Crop. Res. 261:108010. doi: 10.1016/j.fcr.2020.108010
Gol, L., Haraldsson, E. B., and von Korff, M. (2021). Ppd-H1 integrates drought stress signals to control spike development and flowering time in barley. J. Exp. Bot. 72, 122–136. doi: 10.1093/jxb/eraa261
González, F. G., Slafer, G. A., and Miralles, D. J. (2002). Vernalization and photoperiod responses in wheat pre-flowering reproductive phases. Field Crop. Res. 74, 183–195. doi: 10.1016/S0378-4290(01)00210-6
Hemming, M. N., Fieg, S., James Peacock, W., Dennis, E. S., and Trevaskis, B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomics 282, 107–117. doi: 10.1007/s00438-009-0449-3
Hemming, M. N., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2008). Low-temperature and daylength cues are integrated to regulate Flowering Locus T in barley. Plant Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
Hemming, M. N., Walford, S. A., Fieg, S., Dennis, E. S., and Trevaskis, B. (2012). Identification of high-temperature-responsive genes in cereals. Plant Physiol. 158, 1439–1450. doi: 10.1104/pp.111.192013
Hill, C. B., Angessa, T. T., McFawn, L. A., Wong, D., Tibbits, J., Zhang, X. Q., et al. (2019). Hybridisation-based target enrichment of phenology genes to dissect the genetic basis of yield and adaptation in barley. Plant Biotechnol. J. 17, 932–944. doi: 10.1111/pbi.13029
IPCC (2013). Summary For Policymakers. Climate Change 2013: the Physical Science Basis. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press.
Jacott, C. N., and Boden, S. A. (2020). Feeling the heat: developmental and molecular responses of wheat and barley to high ambient temperatures. J. Exp. Bot. 71, 5740–5751. doi: 10.1093/jxb/eraa326
Jamieson, P. D., Brooking, I. R., Semenov, M. A., and Porter, J. R. (1998). Making sense of wheat development: a critique of methodology. Field Crop. Res. 55, 117–127. doi: 10.1016/S0378-4290(97)00072-5
Jones, H., Civá, P., Cockram, J., Leigh, F. J., Smith, L. M. J., Jones, M. K., et al. (2011). Evolutionary history of barley cultivation in Europe revealed by genetic analysis of extant landraces. BMC Evol. Biol. 11:320. doi: 10.1186/1471-2148-11-320
Jones, H., Leigh, F. J., Mackay, I., Bower, M. A., Smith, L. M. J., Charles, M. P., et al. (2008). Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the fertile crescent. Mol. Biol. Evol. 25, 2211–2219. doi: 10.1093/molbev/msn167
Karsai, I., Igartua, E., Casas, A. M., Kiss, T., Soós, V., Balla, K., et al. (2013). Developmental patterns of a large set of barley (Hordeum vulgare) cultivars in response to ambient temperature. Ann. Appl. Biol. 162, 309–323. doi: 10.1111/aab.12023
Karsai, I., Mészáros, K., Láng, L., Hayes, P. M., and Bedo, Z. (2001). Multivariate analysis of traits determining adaptation in cultivated barley. Plant Breed. 120, 217–222. doi: 10.1046/j.1439-0523.2001.00599.x
Karsai, I., Szucs, P., Koszegi, B., Hayes, P. M., Casas, A., Bedo, Z., et al. (2008). Effects of photo and thermo cycles on flowering time in barley: a genetical phenomics approach. J. Exp. Bot. 59, 2707–2715. doi: 10.1093/jxb/ern131
Karsai, I., Szucs, P., Mészáros, K., Filichkina, T., Hayes, P. M., Skinner, J. S., et al. (2005). The Vrn-H2 locus is a major determinant of flowering time in a facultative x winter growth habit barley (Hordeum vulgare L.) mapping population. Theor. Appl. Genet. 110, 1458–1466. doi: 10.1007/s00122-005-1979-7
Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T., and Handa, H. (2009). Molecular and functional characterization of pebp genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353. doi: 10.1104/pp.108.132134
Kiss, T., Dixon, L. E., Soltész, A., Bányai, J., Mayer, M., Balla, K., et al. (2017). Effects of ambient temperature in association with photoperiod on phenology and on the expressions of major plant developmental genes in wheat (Triticum aestivum L.). Plant Cell Environ. 40, 1629–1642. doi: 10.1111/pce.12971
Laurie, D. A., Griffiths, S., Dunford, R. P., Christodoulou, V., Taylor, S. A., Cockram, J., et al. (2004). Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crop. Res. 90, 87–99. doi: 10.1016/j.fcr.2004.07.007
Laws, M. R., Reinheimer, J. L., Coventry, S. J., and Eglinton, J. K. (2010). “Introgression and validation of reproductive frost tolerance,” in Proceedings of the 10th International Barley Genetics Symposium, eds S. Ceccarelli and S. Grando (Alexandria: ICARDA), 222–229.
Lê, S., Josse, J., and Husson, F. (2008). FactoMineR. An R package for multivariate analysis. J. Stat. Softw. 25, 1–18.
Mansour, E., Moustafa, E. S. A., Qabil, N., Abdelsalam, A., Wafa, H. A., El Kenawy, A., et al. (2018). Assessing different barley growth habits under Egyptian conditions for enhancing resilience to climate change. Field Crop. Res. 224, 67–75. doi: 10.1016/j.fcr.2018.04.016
Monteagudo, A., Igartua, E., Contreras-Moreira, B., Gracia, M. P., Ramos, J., Karsai, I., et al. (2019). Fine-tuning of the flowering time control in winter barley: the importance of HvOS2 and HvVRN2 in non-inductive conditions. BMC Plant Biol. 19:113. doi: 10.1186/s12870-019-1727-9
Monteagudo, A., Kiss, T., Mayer, M., Casas, A. M., Igartua, E., and Karsai, I. (2020). Genetic diversity in developmental responses to light spectral quality in barley (Hordeum vulgare L.). BMC Plant Biol. 20:207. doi: 10.1186/s12870-020-02416-1
Mulki, M. A., Bi, X., and von Korff, M. (2018). Flowering locus T3 controls spikelet initiation but not floral development. Plant Physiol. 178, 1170–1186. doi: 10.1104/pp.18.00236
Mulki, M. A., and von Korff, M. (2016). CONSTANS controls floral repression by up-regulating VERNALIZATION2 (VRN-H2) in Barley. Plant Physiol. 170, 325–337. doi: 10.1104/pp.15.01350
Muñoz-Amatriaín, M., Hernandez, J., Herb, D., Baenziger, P. S., Bochard, A. M., Capettini, F., et al. (2020). Perspectives on low temperature tolerance and vernalization sensitivity in barley: prospects for facultative growth habit. Front. Plant Sci. 11:585927. doi: 10.3389/fpls.2020.585927
Nishida, H., Ishihara, D., Ishii, M., Kaneko, T., Kawahigashi, H., Akashi, Y., et al. (2013). Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiol. 163, 804–814. doi: 10.1104/pp.113.222570
Oliver, S. N., Deng, W., Casao, M. C., and Trevaskis, B. (2013). Low temperatures induce rapid changes in chromatin state and transcript levels of the cereal VERNALIZATION1 gene. J. Exp. Bot. 64, 2413–2422. doi: 10.1093/jxb/ert095
Pankin, A., Campoli, C., Dong, X., Kilian, B., Sharma, R., Himmelbach, A., et al. (2014). Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198, 383–396. doi: 10.1534/genetics.114.165613
Prasad, P. V. V., and Djanaguiraman, M. (2014). Response of floret fertility and individual grain weight of wheat to high temperature stress: sensitive stages and thresholds for temperature and duration. Funct. Plant Biol. 41, 1261–1269. doi: 10.1071/FP14061
Russell, J., Mascher, M., Dawson, I. K., Kyriakidis, S., Calixto, C., Freund, F., et al. (2016). Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48, 1024–1030. doi: 10.1038/ng.3612
Shaw, L. M., Li, C., Woods, D. P., Alvarez, M. A., Lin, H., Lau, M. Y., et al. (2020). Epistatic interactions between PHOTOPERIOD1, CONSTANS1 and CONSTANS2 modulate the photoperiodic response in wheat. PLoS Genet. 16:e1008812. doi: 10.1371/journal.pgen.1008812
Slafer, G. A., and Rawson, H. M. (1994). Sensitivity of wheat phasic development to major environmental factors: a reexamination of some assumptions made by physiologists and modellers. Aust. J. Plant Physiol. 21, 393–426. doi: 10.1071/PP9940393
Trevaskis, B., Hemming, M. N., Peacock, W. J., and Dennis, E. S. (2006). HvVRN2 Responds to Daylength, whereas HvVRN1 Is Regulated by Vernalization and Developmental Status. Plant Physiol. 140, 1397–1405. doi: 10.1104/pp.105.073486.1
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U. S. A. 100, 13099–13104. doi: 10.1073/pnas.1635053100
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The Pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–12. doi: 10.1186/gb-2002-3-7-research0034
von Zitzewitz, J., Szûcs, P., Dubcovsky, J., Yan, L., Francia, E., Pecchioni, N., et al. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59, 449–467. doi: 10.1007/s11103-005-0351-2
Wiegmann, M., Maurer, A., Pham, A., March, T. J., Al-Abdallat, A., Thomas, W. T. B., et al. (2019). Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci. Rep. 9:6397. doi: 10.1038/s41598-019-42673-1
Wollenweber, B., Porter, J. R., and Schellberg, J. (2003). Lack of interaction between extreme high-temperature events at vegetative and reproductive growth stages in wheat. J. Agron. Crop Sci. 189, 142–150. doi: 10.1046/j.1439-037X.2003.00025.x
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U. S. A. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The Wheat VRN2 Gene Is a Flowering Repressor Down-Regulated by Vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U. S. A. 100, 6263–6268. doi: 10.1073/pnas.0937399100




